Home | Duong Vu's personal page
It’s been a while since I last posted here. I’ve remained active on Twitter, though balancing multiple platforms can be a bit challenging and time-consuming. 😊
From now on, please follow our work on our MycoAI page. There, you’ll find all the software and tools we develop and maintain, along with our publications and updates on our activities.

This week, we had the kickoff meeting for the ‘AID: Artificial Intelligence for DNA barcode identification’ project, which was recently granted by
the Netherlands eScience Center. This is a joint project between the Westerdijk Institute
and ARISE. A short description of the project available
the eScience Center’s research-software-directory.
Please follow the project’s updates at our GitHub page: https://github.com/MycoAI.
While creating a logo for MycoAI using Generative AI, I’ve also created a new avatar for my GitHub page: https://github.com/vuthuyduong.
That’s really me on the image, working hard on finding the best AI models for identifying fungal biodiversity :)
The webinterface dnabarcoder.org of dnabarcoder, developed by my student, Ruby van der Holst, from Hogeschool Leiden during her internship at our institute, is ready to use. So proud of her work!

Thank you BioSB2023 for giving me the opportunity to present the Westerdijk fungal data resources for bioinformatics and biology. It was great to see many familiar faces here again, as well as the new ones working in Bioinformatics around the NL and other places of the world.

I am honored to become a new member of the UNITE community, and very happy to see that the first UNITE General Assembly was hosted at the Westerdijk Institute.

It was a great honor for me to present the work from the Westerdijk Institute on identifying fungal biodiversity from environmental eDNA samples in the ARISE project to Minister Dijkgraaf.

Wow Today’s issue of The Washington Post features a press release of our recent study
How, not if, is the question mycologists should be asking about DNA-based typification, led by Henrik R. Nilson discussing about DNA-based descriptions (typifications) of fungal species.
This study has been all over the media in the last few weeks, but appearing on The Washington Post is really special…
I am so excitied to see this paper Comparing genomic variant identification protocols for Candida auris, led by Christina A. Cuomo and Anastasia P. Litvintseva, being published.
With Ferry Hagen, we contributed a pipeline to this study that is available at my github: https://github.com/vuthuyduong/SNPanalysis. This pipeline can be used for Illumina, PacBio, and Nanopore reads.

It was a great honor to receive this distinguished delegation from the Vietnam Academy of Sciences and Technology (VAST), led by Prof Chu Hoang Ha,
Vice-President of VAST to visit the Westerdijk Institute yesterday. Looking forward to our collaboration!
So happy to see that an entry
in the research software directory has been created for our recently funded Small-Scale Initiatives Machine Learning project by the Netherlands eScience Center

I would like to share with you a great news: Together with Gerard Verkleij, we won the Dutch Data Prize 2022 in the Life Sciences category for the dataset released in the paper Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation.

Up and down the mountains in South Africa looking for fungi, feeling like I am an ecologist in making :).
A heartfelt gratitude to the Johanna Westerdijkfonds and the EU-funded project Mycobiomics No. 101008129 for making my dreams a reality! Their support has been invaluable in furthering our research endeavors in this region.

I am grateful for the opportunity to present our research at the SASSP2022 conference on fungal species identification, particularly the generation of a comprehensive barcode dataset as part of the DNA barcoding project at the Westerdijk Fungal Biodiversity Institute. This extensive dataset is a significant achievement and a valuable resource for fungal biodiversity studies. I would like to thank the EU H2020-MSCA-RISE project Mycobiomics No. 101008129 and the Johanna Westerdijkfonds for their generous sponsorship, which made it possible for me to participate in the conference and conduct fieldwork trips in South Africa.
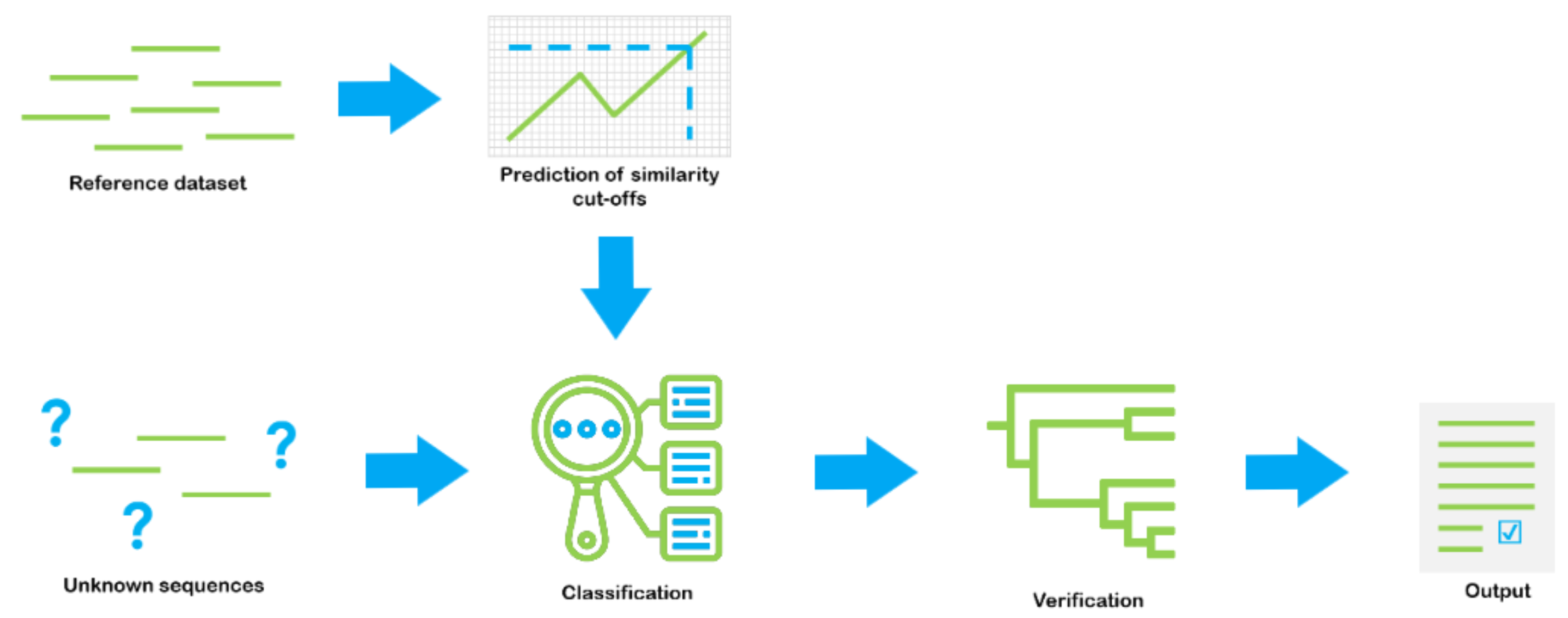
Our paper Dnabarcoder: An open-source software package for analysing and predicting DNA sequence similarity cutoffs for fungal sequence identification is out!
“Metabarcoding loses significant resolution and scientific explanatory power by relying on a single similarity cut-off for taxonomic assignment”.
Dnabarcoder can be used to calculate similarity cutoffs for sequence identification for different clades of fungi and other organisms. The source code of the dnabarcoder is available at my github page: https://github.com/vuthuyduong/dnabarcoder.
Citation
Duong Vu, R. Henrik Nilsson, Gerard J.M. Verkley (2022). dnabarcoder: an open-source software package for analyzing and predicting DNA sequence similarity cut-offs for fungal sequence identification. Molecular Ecology Resources. https://doi.org/10.1111/1755-0998.13651

Such a great conference ICDBB2022! Thank you for providing us with the opportunity to present our newly developed tool,
dnabarcoder,
which enables accurate identification of fungal species from environmental samples using the eDNA approach.
This project is a collaboration between Gerard Verkleij from the Westerdijk Institute and Henrik Nilsson from the UNITE community.

Currently, only approximately 150,000 of the estimated 6 million fungal species have been described. Describing all fungal species through culturing alone is estimated to take us 2,000 years. In a recent study
Fungal taxonomy and sequence-based nomenclature
led by Robert Lücking and Conrad L. Schoch, a sequence-based approach for the nomenclature of taxa known only from sequences was discussed. This approach aims to facilitate consistent reporting and communication in the literature and public sequence repositories. You can find an introduction about the study in this blog post. The poster image in the blog post showing the molecular fungal “universe” was derived from figure 3 in the paper. The image was created using our tool fMLC (Windows version), published in the paper fMLC: fast multi-level clustering and visualization of large molecular datasets.
I am very happy to share with you the excellent news that our paper titled Convolutional Neural Networks Improve Fungal Classification has been made publicly available.
This is the first time we have applied the deep learning approach with two models: Convolutional Neural Network (CNN) and a Deep Belief Network (DBN) for fungal classification. We have compared the results with traditional methods such as BLAST classification and the most accurate machine learning-based Ribosomal Database Project (RDP). Our findings show that the deep learning approach outperformed the traditional methods in cases where we had enough reference sequences in the training dataset. However, when classifying a dataset that is not present in the training dataset, BLAST could reveal the most sequences. The source code of the CNN and DBN classifiers as well as the comparion can be found at my github page: https://github.com/vuthuyduong/fungiclassifiers.
The identification of fungi is challenging as fungi have simple body plans with often morphologically and ecologically obscure structures, compared with plants and animals.
In this study Unambiguous identification of fungi: where do we stand and how accurate and precise is fungal DNA barcoding?, led by
Robert Lücking Conrad L. Schoch, we discussed accuracy, precision, and challenges of current approaches for fungal identification.
A conceptual framework for the identification of fungi, encouraging the approach of integrative (polyphasic) taxonomy for species delimitation, i.e. the combination of genealogy (phylogeny),
phenotype (including autecology), and reproductive biology (when feasible), was provided.